Unlocking Protein-Drug Interaction Secrets for Safer Drugs
Understanding protein-drug interactions is crucial in the development of new therapeutics. By exploring the complex network of drug-protein relationships, researchers gain insights into drug efficacy and safety. The premise is simple: drugs that share more target proteins have a higher likelihood of similarity in their effects and interactions.
The aim is to refine predictions and enhance drug discovery processes. Researchers leverage advanced methods like the drug-protein interaction network space to establish similarity matrices, which are instrumental in predicting potential interactions. This not only streamlines the drug development pipeline but also introduces a higher degree of precision in targeting specific proteins.
Importance of Protein-Drug Interactions in Therapeutics
Understanding protein-drug interactions is not just an academic exercise; it’s at the core of developing effective and safe therapeutics. These interactions dictate how a drug works, its efficacy, and potential side effects.
- By exploring protein-drug interactions, researchers are able to map out how drugs influence specific proteins. This insight is precious—it’s the difference between a shotgun approach and a precision strike in therapeutics.
With the advent of systems biology, protein-drug interaction research has surged ahead. Researchers now have the capacity to comprehensively analyze drug effects at the proteomic level, identifying both target and off-target interactions. This granular level of detail helps us understand the drug’s therapeutic effects and its potential to cause unwanted side effects.
- Critically, researchers can investigate the pleiotropic activities of drugs, which have effects on multiple different proteins or biological pathways. This opens doors to repurposing established drugs for new therapeutic uses, which can be significantly more time and cost-effective than developing new drugs from scratch.
The advanced methodologies for mapping drug-protein interaction networks are key to demystifying the complex relationships between drugs and the proteins they interact with. By establishing similarity matrices, researchers can predict how a drug will interact with its target protein – an essential step for the precise tailoring of treatments.
Moreover, as researchers constantly seek to improve our understanding, we’re looking into the development of powerful statistical models that can accurately detect these interactions. The data derived from these models are invaluable for deciphering the biological mechanisms at play and thus for the rational design of new medication.
Every new insight gained in this field pushes the frontiers of pharmaceutics, enabling us to design drugs that are both potent and safe. It’s our mission to continue to elucidate these associations, advancing therapeutics, and ultimately delivering better health outcomes. Through our efforts, protein-drug interaction knowledge stands to become an increasingly critical piece of the puzzle in modern medicine.
Exploring the Complex Network of Drug-Protein Relationships
Drug discovery and development hinge critically on our understanding of drug-protein interactions. These relationships are at the heart of therapeutic efficacy and safety. With protein-drug interaction forming the cornerstone of drug design, it’s imperative that we delve into the underlying networks that govern these interactions.
The landscape of drug-protein interactions is immense and intricate. High-throughput computational methods have revolutionized our capacity to parse through this complexity, revealing vast networks of relationships. Hundreds of FDA-approved drugs and candidates in late-stage clinical trials present us with a repository of possibilities for drug repositioning. This strategy not only saves significant resources but also accelerates the availability of treatments.
| Molecule Type | Therapeutic Applications | Analysis Strategy
|
| Small Molecules | Anti-Cancer Therapy | Binding Hot Spots Identification |
| Peptides | Anti-Cancer Therapy | Chemical Similarity Incorporation |
By focusing on these systems, we explore a multitude of strategies, each aimed at enhancing drug efficacy and minimizing adverse reactions. Identifying binding hot spots and integrating chemical similarity data are just two of the approaches we leverage to tackle the challenge of protein-protein interaction targeting, especially in complex fields like anticancer therapy.
Interactions within ion channel networks exemplify the level of detail accessible through contemporary analysis. Research points out the completeness of these networks by measuring the proportion of unreachable paths—illuminating the profound connectivity that exists between drugs and their protein targets. By employing advanced computational protocols, a clearer understanding of drug efficiency and potential new applications is attainable.
Yet, alongside these developments, there’s a noted trend of “me-too” drugs—medications that intersect with already established protein targets. While this suggests a high level of redundancy, it also underscores the necessity for innovative approaches to identify novel drug-protein interactions.
Our exploration is more than just a pursuit of new knowledge. It’s a race towards better health outcomes—an amalgamation of science, technology, and the relentless quest to refine and personalize therapeutics. We’re not just mapping an intricate network; we’re paving the way for the next breakthroughs in drug discovery and patient care.
Understanding Drug Efficacy and Safety through Protein-Drug Interactions
Delving into the arena of protein-drug interaction is akin to unlocking a puzzle essential to drug efficacy and safety. Grasping the ways drugs affect protein behavior can offer vital clues about their therapeutic potential and side effects. Here’s what we’ve gathered about how these interactions impact drug development and treatment approaches.
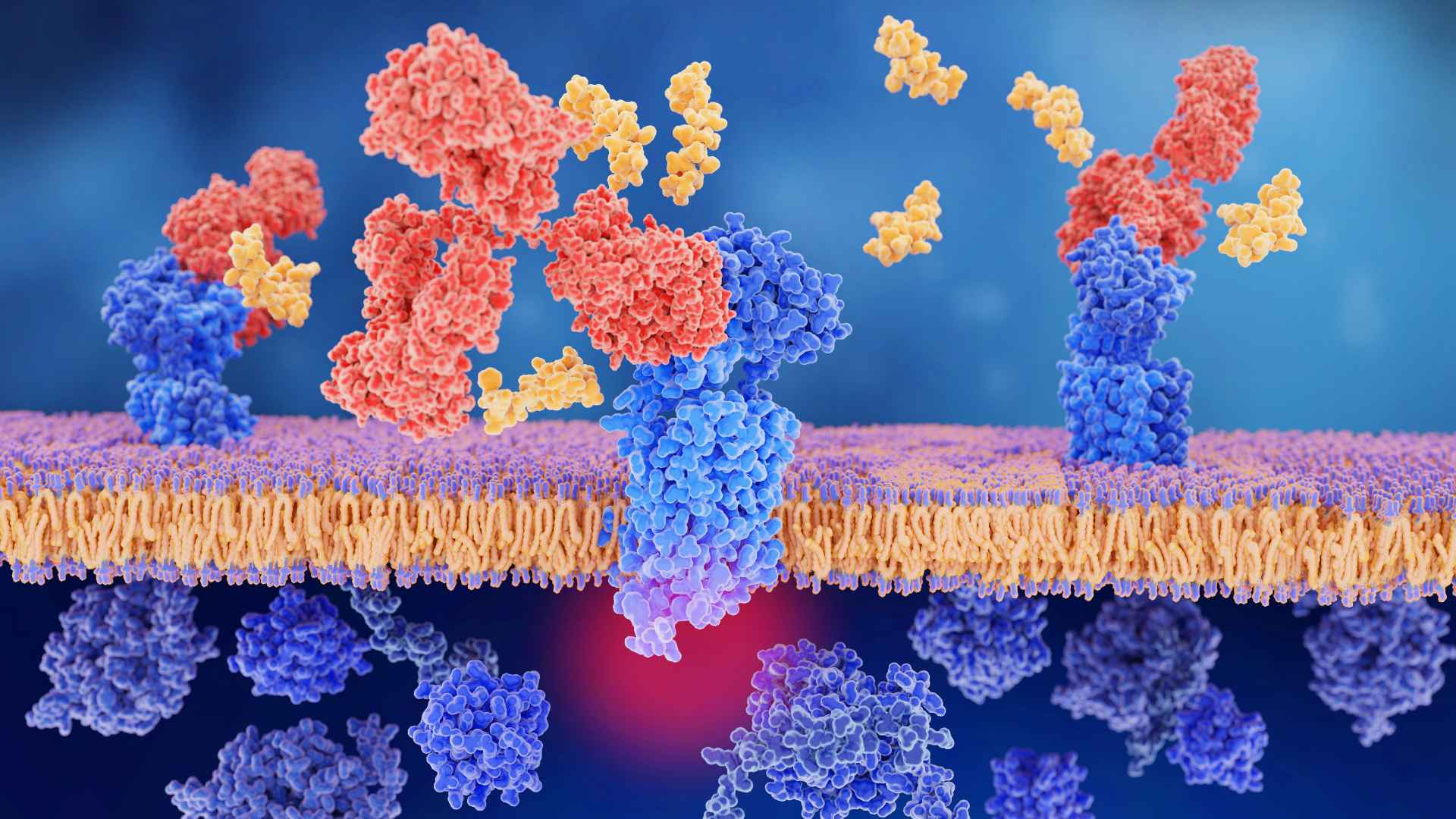
Drug-Efficacy: At the core of our research, it’s the interplay between drugs and proteins that dictates how effectively a drug performs its intended function. Proteins, often being the target of many drugs, serve as the machinery within the body that drugs must engage with to exert their effects. By binding to proteins, drugs can either activate or inhibit their functions, leading to the intended therapeutic outcomes.
On the flip side, identifying off-target effects is crucial in understanding the safety profile of drugs. When drugs interact with proteins other than their primary targets, they can trigger a cascade of unintended reactions, sometimes beneficial, but often leading to adverse effects. This understanding helps us mitigate risks and tailor drug dosages more precisely.
Here are some statistics underscoring the significance of this relationship:
| Variable | Value
|
| Number of FDA-approved drugs | Thousands |
| Drug development time | Years |
| Drug repositioning savings | Time & Cost |
The recognition that repurposing FDA-approved drugs for new therapeutic uses isn’t just convenient—it’s a strategic move guided by our insight into protein-drug interactions. It’s a method that saves both time and costs, a critical consideration given the intense regulatory environment and the hefty price tag of traditional drug discovery.
We’re committed to advancing our analytics to map out these complex interaction networks. By quantitatively assessing drug affinity and specificity, we set the stage for refining drug design, improving therapeutic indices, and predictive drug repositioning. It’s a quest to ensure that every drug released not only achieves its desired effect but does so with the utmost safety.
The Relationship between Shared Target Proteins and Drug Effects
Understanding protein-drug interactions is pivotal in grasping how medicinal compounds exert their effects on biological systems. Drugs achieve their therapeutic outcomes by binding to specific proteins within the body, influencing the protein’s activity. This can result in either an increase or decrease in normal protein function, which correlates with the drug’s intended benefits or potential side effects.
Proteins often serve as the key interplayers in this dynamic, with some acting as the primary targets for multiple drugs. Recognizing these interactions is not just about identifying a drug’s effectiveness, but also about predicting its side effects and drug-drug interactions that may occur due to shared protein targets. This knowledge empowers us to optimize treatment regimens for patients and tailor drug development more precisely toward desired therapeutic effects.
The impact of drugs on shared target proteins can be extensive, influencing various pathways that are critical for maintaining homeostasis. When multiple drugs compete for the same target protein, they might dampen each other’s efficacy or intensity side effects. It’s crucial for us to predict these outcomes through protein-drug interaction studies, as they can provide insights into the drug’s pharmacodynamics and pharmacokinetics.
Through advanced analytical methods, like the mass spectrometry-based proteomics approach, we’ve begun to uncover complex networks of protein-drug interactions. Such analysis can indicate not only the direct targets that drugs interact with but also the subtle off-target effects that might go unnoticed without this deep dive into protein behavior.
Our exploration into these intricate networks does not just stop with understanding. Armed with data concerning how drugs and proteins behave within these networks, we’re able to influence drug design and repurposing strategies more effectively. As we unravel more about the shared path’s drugs take by targeting the same proteins, we can better strategize their use to enhance therapeutic indices and mitigate risks.
In doing so, we extend our comprehension beyond a drug’s primary effects to appreciate the full spectrum of its influence on the proteome. This awareness carries immense value for repurposing existing FDA-approved drugs, which could catalyze the emergence of novel therapies. Through this, we seamlessly integrate our growing knowledge of protein-drug interactions into advancing therapeutic strategies that prioritize both efficacy and safety.
Refining Predictions and Enhancing Drug Discovery Processes
In our journey to advance drug discovery, protein-drug interaction plays a pivotal role. As we better understand these interactions, we’re able to refine predictions which in turn amplifies the efficiency of the entire process. But how does one streamline the prediction of protein-drug interaction?
Our focus has shifted towards the optimization of computational models. Leveraging the vast databases of FDA-approved drugs, our methods hone in on potential new applications. This approach not only saves time but also resources ordinarily sunk into de novo drug development.
- Drug repositioning has emerged as a key strategy to repurpose existing drugs for new therapeutic uses.
- In silico prediction models expedite identifying promising drug-protein pairs.
But why stop at predictions? One of our objectives is to corroborate these predictions with laboratory results. We’ve integrated various high-throughput screening tools, with a significant nod to automation in accelerating both drug discovery and validation stages.
- Automated pipelines, like those enabled by KNIME, assist in seamless data processing.
- Advanced statistical methods detect drug-protein interactions more effectively, leading to targeted drug design.
In all these efforts, we must bear in mind the biological mechanisms at play. It’s not just about which drugs interact with which proteins, but unraveling the complexity of these interactions to design better therapeutics. With computational power on our side, the scale of data we can analyze is unprecedented, allowing us to draw more accurate connections between molecular structures and their interactions.
Ensuring the therapeutic efficacy of drugs is as much about the ingenuity of prediction as it is about the thoroughness of subsequent testing. By honing our predictive capabilities and combining them with robust high-throughput techniques, we’re ever enhancing the pace and precision of drug discovery. It’s a continuing effort to align efficiency with outcome, where success is measured not only in theoretical potential but in real-world therapeutic benefits.
Leveraging Advanced Methods for Predicting Potential Interactions
In our quest to enhance drug discovery, we’ve made significant strides in predicting potential protein-drug interactions. Such predictions are crucial for identifying not only primary therapeutic targets but also unintended off-target effects that could lead to side effects or contribute to drug repurposing strategies.
One of the most promising techniques we’re utilizing is mass spectrometry-based proteomics. This advanced analytical method allows us to dissect the intricate networks that define protein-drug interactions. By utilizing this technology, we’re able to illuminate both direct targets and subtle off-target effects that may not be evident through traditional methods.
- Mass spectrometry-based proteomics
- Illuminating direct and off-target effects
The approach includes a manifold regularization semi-supervised learning method, which uses both labeled and unlabeled data. This machine learning strategy effectively overcomes the challenge of limited known interactions by harnessing the wealth of unexplored data, often yielding more superior predictions than methods relying on labeled data alone.
- Semi-supervised learning
- Harnessing labeled and unlabeled data
Moreover, this method doesn’t work in isolation. It integrates heterogeneous biological data, such as chemical structures and genomic sequences, alongside known interaction networks. By doing so, we leverage all available information, improving the accuracy and reliability of our predictions.
- Integration of diverse biological data
- Improved prediction accuracy
Table: Summary of Our Predictive Methods Benefits
| Method | Benefits
|
| Mass Spectrometry-Based Proteomics | Identifies direct and off-targets |
| Semi-Supervised Learning | Utilizes both labeled and unlabeled data |
| Integrating Biological Data | Enhances accuracy and reliability |
We’re now applying these methods to predict interactions within specific datasets, namely enzymes, ion channels, GPCRs, and nuclear receptors. Our prediction results have shown considerable promise, with some findings confirming the latest pharmacological data.
As we continue to hone in on these advanced techniques, our understanding of protein-drug interactions deepens, paving the way for future breakthroughs in drug discovery and development. Through this journey, we remain committed to revealing the complex nature of these interactions, always with the goal of maximizing therapeutic efficacy while minimizing risks.
Streamlining the Drug Development Pipeline through Protein-Drug Interactions
The landscape of pharmaceutical R&D has evolved to appreciate that protein-drug interactions (PDIs) are foundational in the development of efficacious treatments. Recognizing the potential application of FDA-approved drugs in new therapeutic categories—an approach known as drug repositioning—relies on understanding these crucial interactions. The quest for efficient drug discovery leads us through a maze of regulatory prerequisites, monitoring drug toxicity, and ensuring therapeutic efficacy. Yet, the drive for innovation persists.
Automation and cutting-edge techniques are pillars in uncovering the myriad of interactions that govern drug efficacy. With advanced computational models and high-throughput screening tools at our disposal, we are now poised to redefine the paragon of drug discovery. By integrating these quantitative systems, we significantly reduce time-to-market for new therapies and enhance our understanding of the PDIs.
One cannot overstate the importance of in vitro monitoring of PDIs during drug discovery. These interactions underpin the therapeutic effects of drugs. Our commitment to honing in on these physical interactions between small molecules and proteins leads us to leverage high-fidelity, high-throughput screening methods. Subsequently, we surmount challenges associated with traditional biophysical approaches—labelling issues, accuracy reduction due to immobilization, and the low sensitivity detection limitations for small-molecule drugs.
As we streamline the processes of structure-activity relationship (SAR) and mode-of-action (MOA) studies, we usher in a new paradigm. Drug screening, no longer shackled by the constraints of expensive and low-sensitivity methods, paves the way for unprecedented progress. The meticulously reconstructed drug-protein interaction networks inform us not only about potential direct targets but also reveal subtle off-target effects that might influence drug repurposing strategies. It is through these insights that we can truly tailor therapies that align with our goal of amplifying therapeutic indices while judiciously curtailing risks.
Our dedication to refining these predictive efforts enables us to thrust forward, making strides in both efficiency and outcome. We do so by capitalizing on statistical methods designed for the rigors of drug development while staying rooted in the biological mechanisms that drive the success of current and future therapeutics.
Introducing Precision in Targeting Specific Proteins
As we delve deeper into the intricate web of protein-drug interactions, our aim is to forge a path toward precision medicine that can identify specific protein targets with unremitting accuracy. We understand that achieving the desired therapeutic effect hinges on accurately targeting these proteins. With the adoption of mass spectrometry-based proteomics’ services, we’re on the cusp of a breakthrough, pinpointing drug interactions with unprecedented precision.
This advanced analytical method has the potential to revolutionize how we identify the protein targets of drug molecules. Through thermodynamic measurements of protein-folding reactions within complex biological mixtures, we can detect protein-drug interactions that would otherwise remain hidden. The binding affinity, such as the one between cyclosporin A (CsA) and Cyclophilin A with a dissociation constant (Kd) of 70nM, exemplifies the method’s rigor.
Discovering protein targets, both direct and indirect, offers a nuanced understanding of a drug’s behavior. For instance, our identification of UDP-glucose4-epimerase as an indirect target during our proof of principle work with CsA exemplifies this method’s capability to discern subtle protein-drug interactions that mediate side effects or therapeutic benefits.
| Known Protein Targets of CsA | Type of Interaction | Dissociation Constant (Kd)
|
| Cyclophilin A | Direct | 70nM |
| UDP-glucose4-epimerase | Indirect | Not Specified |
Notably, beyond targeting proteins implicated in disease, our methods have implications for understanding the broader protein-folding environment. This encompasses a range of orphan diseases and the aging process, where protein misfolding plays a significant role. By leveraging our technology in such applications, we offer a beacon of hope, anticipating that early predictions of protein-drug interactions will lead to more precise therapeutic interventions with fewer side effects.
Conclusion
We’ve explored the intricate landscape of protein-drug interactions and their pivotal role in shaping the future of pharmacology. Our journey through the complexities of these interactions reaffirms their significance in drug discovery and patient care. By harnessing the power of cutting-edge technologies like mass spectrometry-based proteomics and semi-supervised learning, we’re poised to unlock a new era of precision medicine. As we continue to refine our methods and deepen our understanding, we’re committed to advancing therapeutics that are not only effective but also safe.
The path ahead is clear: to deliver on the promise of tailored treatments that address the unique needs of individuals while mitigating the risks associated with drug therapy. Together we’re forging ahead to a future where every patient can benefit from the meticulous craft of drug design and the profound insights of protein-drug interaction research.